covid_proportion_estimation
Test Positivity Rate Distribution Dashboard 
The goal of Test Positivity Rate Distribution Dashboard is to estimate of the distribution of COVID-19 test positivity rates in communities.
Installation
You can install the development version from GitHub with:
install.packages("devtools")
devtools::install_github("alexanderjwhite/covid_proportion_estimation")
Access App
There are two ways to access the app:
- https://alexanderjwhite.shinyapps.io/covid_proportion_estimation/
- from the package (see below)
run_app()
Instructions
Welcome to the dashboard of the distribution of test positivity rate. This application is designed to estimate the distribution of test positivity rate among local communities. As an example, a sample of the COVID-19 testing data in each zip code collected by the city of Chicago from 8/30/2020 to 9/5/2020 has been pre-loaded. In the View Data section, the loaded data are displayed with each row representing data from a zip code. The Results section displays the estimates of the proportion of zip code areas with a test positivity rate below various upper thresholds. The Proportion Between Lower and Upper Thresholds section displays the proportion of zip code areas with a test positivity rate between arbitrary lower threshold and upper threshold.
-
Format your data as shown in the image below. The first row should contain the column names. Each row in the column named tests should contain the number of tests performed. Each row in the column named cases should contain the number of positive test results. Acceptable formats include .csv, .xlsx, and .xls.
-
Navigate to the Upload tab.
-
Select Browse and upload your file.
-
A table of default positive rate proportion will populate in the Results table. Estimate the positive rate proportion for a custom range by adjusting the sliding bar and selecting Update.
Background
The distribution of COVID-19 test positivity rate in communities can be
used to guide public health and policy responses. Specifically, the
proportion of local communities (e.g. zip code areas) with a test
positivity rate below an upper threshold (or above a lower threshold)
offers important insight on the disease control within a city or county.
For instance, if 10 out of 50 zip code areas in a city have a test
positivity rate below an upper threshold of 5% then the naïve estimate
of the proportion is
. The naïve estimate, however, ignores the margin
of error of the test positivity rate in each zip code, which could lead
to either over- or under-estimate of the proportion. This online
application uses a statistical method called Bayesian deconvolution to
generate a more accurate estimate.
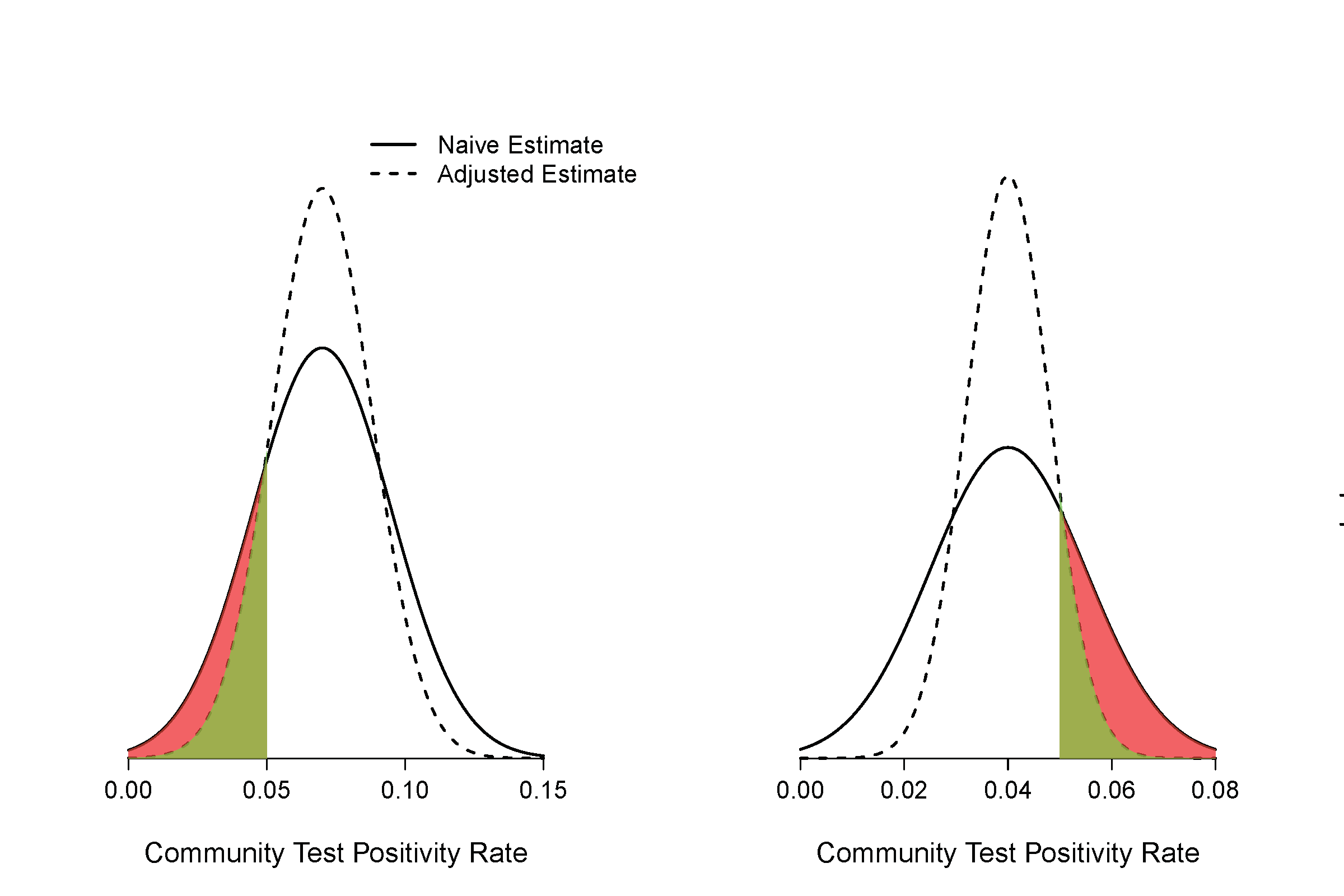
Figure 1
Illustration on how the native calculation can result in overly-optimistic or overly-pessimistic estimate of the proportion of community-level test positivity rate below a threshold.
Legend
The figure on the left side shows one scenario where the naïve estimate will overestimate the proportion of community level test positivity rate of 5% or less as compared with estimate that accounts for the margin of errors (adjusted estimate). The red area indicates the magnitude of over-optimism. The figure on the right side shows one scenario where the naïve estimate will overestimate the proportion of community level test positivity rate of 5% or more. The red area indicates the magnitude of over-pessimism.
Technical Details
Suppose there are
communities. Let
and
be the
number of tests and cases in a given period of time for community
. The naïve estimate of the test positivity rate
for community
,
, has a margin of error in estimating the true
test positivity rate
. Here the true test positivity rate refers to the rate that would
have been calculated had we been able to test all eligible subjects in
the community. The margin of error associated with
in estimating
can be estimated
by
.
The naïve estimate of the proportion of communities with test positivity
rate below threshold is
where is the indicatory function. Because of the margin of errors
associated with
could be a biased estimate of the true
proportion
A solution that accounts for the margin of errors is the empirical Bayes
deconvolution method. In this approach, the distribution induced by the
rates , is assumed to be a natural cubic spline
with knots at equally spaced quantiles and can be estimated through
either maximum likelihood or regularized maximum likelihood method using
data
. We used maximum likelihood method with
Bayesian Information Criteria (BIC) as the tool to select model degrees
of freedom up to 5. Once we obtain an estimate
, we can estimate
by
can be a much more accurate estimate of
than
.
References
- Efron B. Empirical Bayes deconvolution estimates. Biometrika. 2016;103(1):1-20.
- Narasimhan B, Efron B. deconvolveR: A G-Modeling Program for Deconvolution and Empirical Bayes Estimation. Journal of Statistical Software. 2020;94(11).
